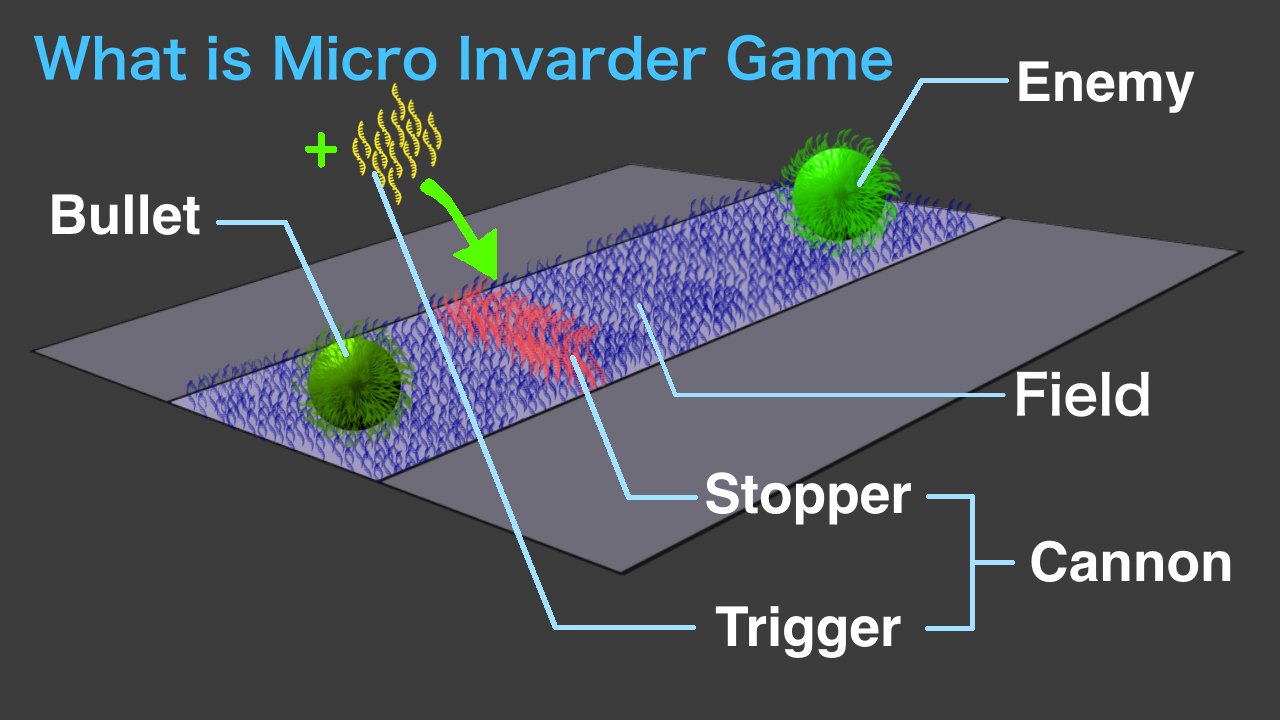
The micro invader game is recreating "Space Invader" in micro-sized form using bio-molecules.
"Space Invader" is a game where players shoot down enemies attacking from space using bullets. Our micro invader game consists of three main components imitating those.
The first component is Enemy, which is made up of liposomes and nucleic acid strands. Enemies fluoresce in green.
The second component is Bullet, which is made of silica beads and nucleic acid strands.
The last component is Cannon, which is a mechanism whereby the Bullet is fired by the player's manipulation. Strand displacement reaction of nucleic acids is used.
These parts are placed on a field designed using avidin-biotin protein and nucleic acid strands (call it Base strand )on a glass surface.
Base strands have complementary sequence to strands on Bullet surface and on the Enemy. This allows Bullet and the enemy to stay on the Field.
Bullets are placed at end of the Field. And at the other end of it, Enemy is placed.
(The nucleotide sequence of the part where the Enemy is placed has a complementary sequence to the nucleotide sequence on the Enemy.)
●Enemy Design
●Making Enemy
○Making Liposome
○Assembling Enemy
●Bullet Design
●Field Design
●Simulating Bullet Dynamics
●Cannon Design
○Design Strands Sequence
●Simulating Cannon Dynamics
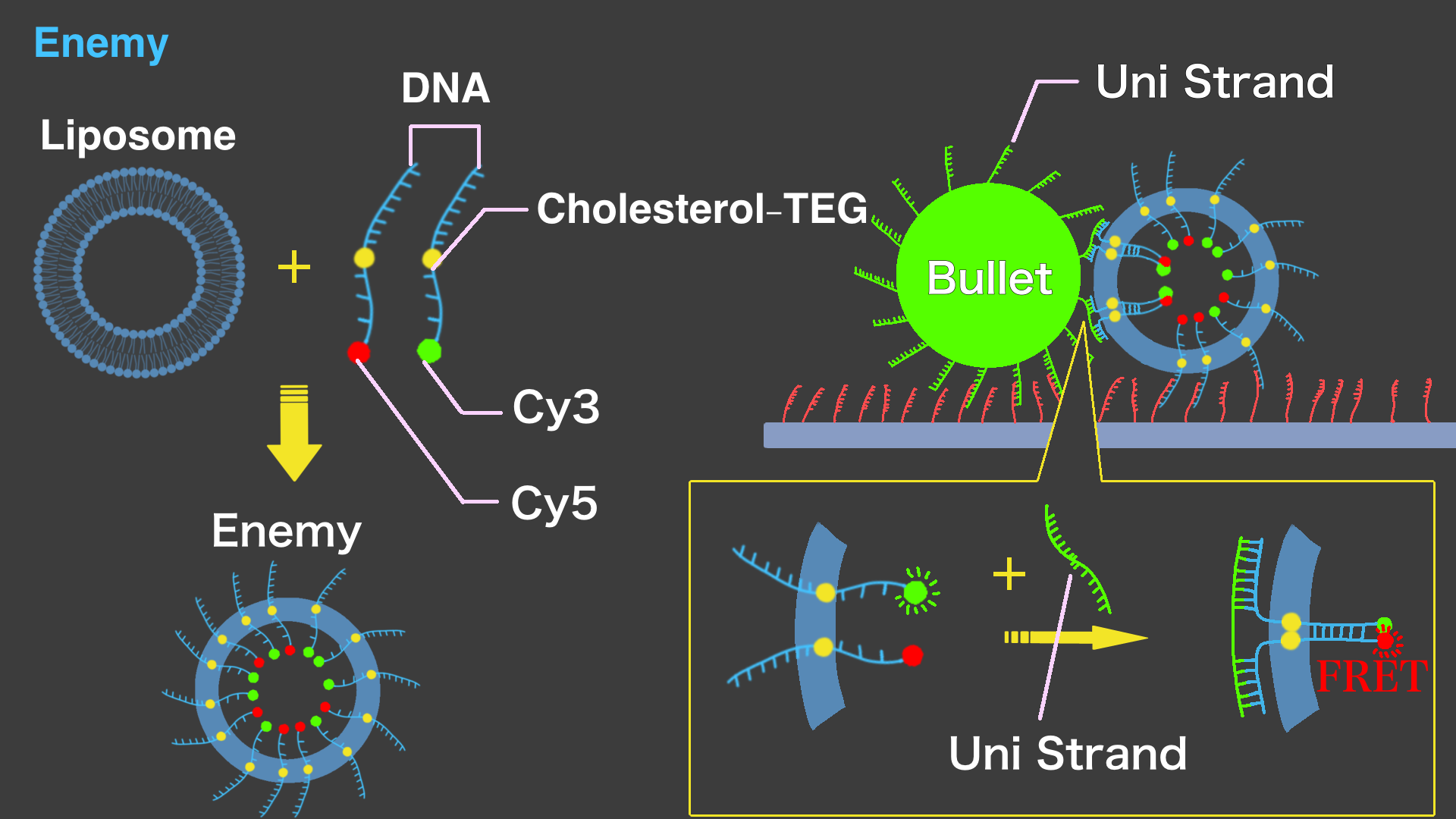
The basic structure of Enemy is a liposome. Enemy's liposome has two types of DNA strands with cholesterol modifications in the center, penetrating the membrane. The outer parts of the DNA strands in Enemy liposome have a complementary sequence to the DNA strand on the surface of Bullet and hybridizes. The inner ends of each DNA strand are modified with Cy3 and Cy5, respectively. When the DNA strand on Bullet and the two DNA strands on Enemy hybridize, the DNA strands on Enemy come closer to each other. This causes FRET (Fluorescence Resonance Energy Transfer) between the fluorescent substances, changing the color of the enemy from green to red. This color change represents the defeat of Enemy in the "Space Invader".
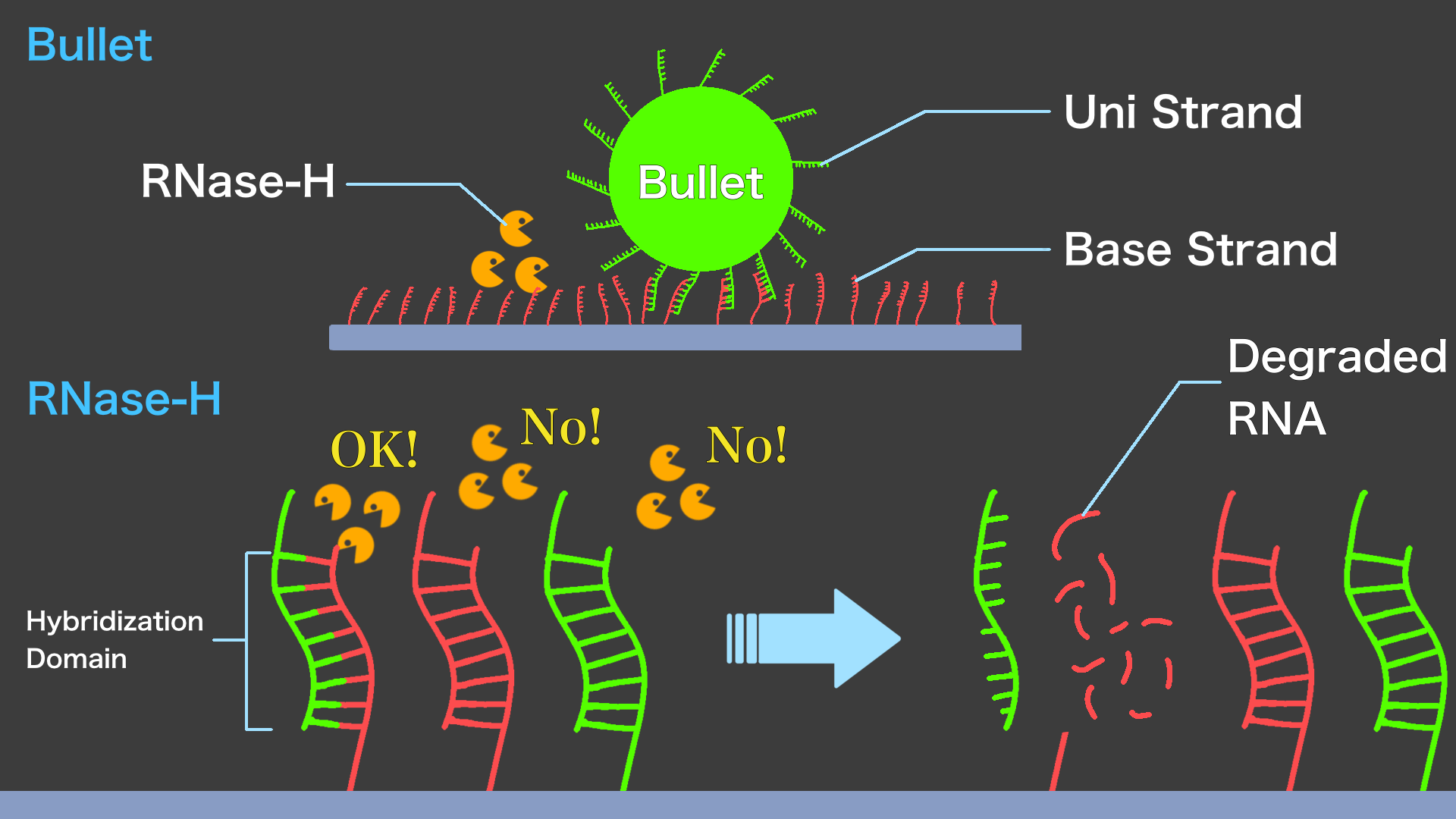
Bullet consist of DNA strands on the surface of a silica beads that are complementary to Base strand and on Enemy. Bullet is anchored to the Field by hybridization with Base strands. To run Bullet towards Enemy, RNase-H is used; RNase-H specifically hydrolyzes the RNA in the DNA/RNA double-stranded structure. This enzyme hydrolyzes Base strand (RNA) hybridizing with DNA on Bullet surface. Bullets that have lost their foothold can move on the Field by rolling and hybridize with new Base strands on the Field.
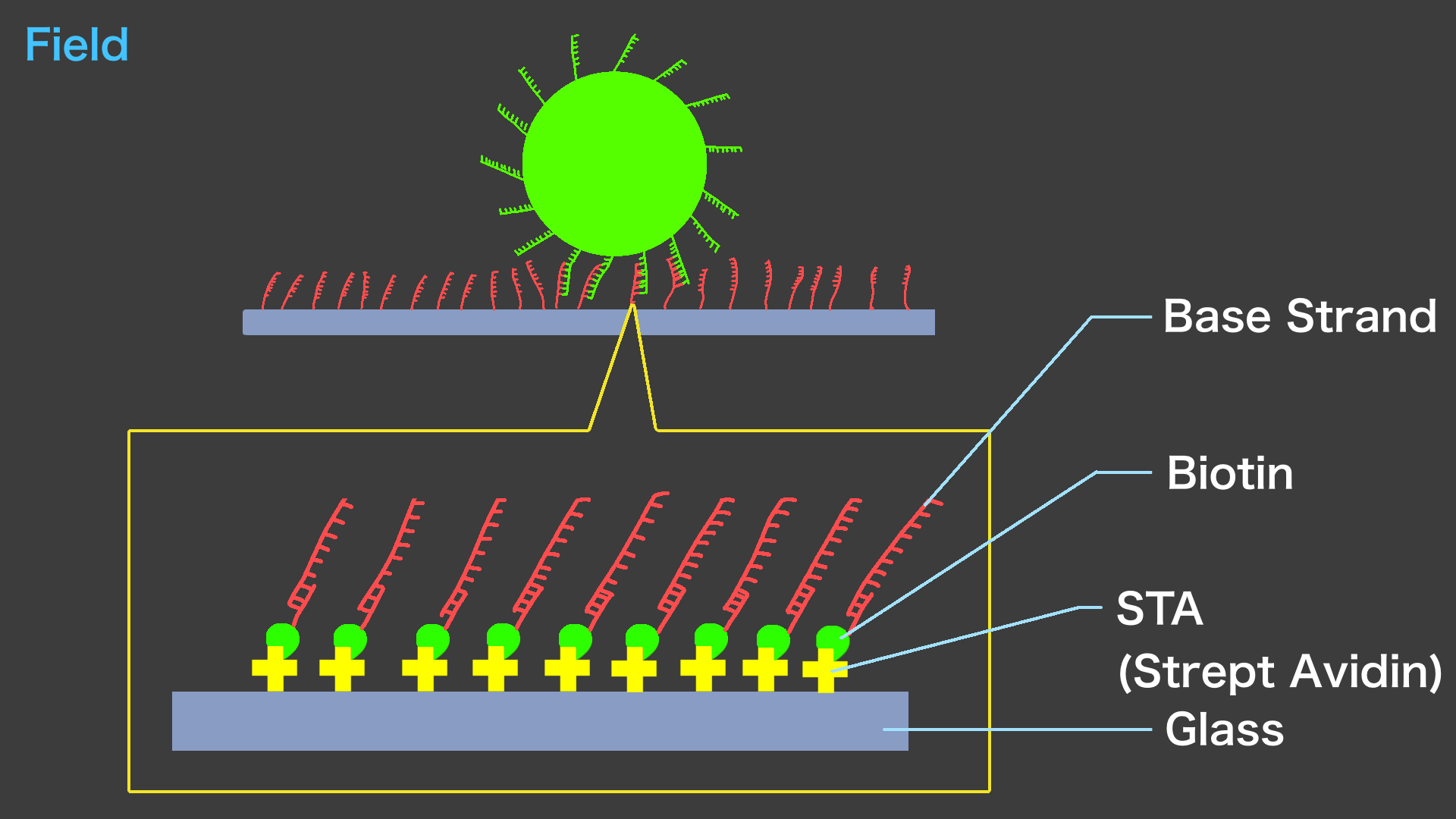
The Field is Base strand adhered to a glass surface. A biosensor is used to adhere the STA to the glass surface. After that, Base strand modified with biotin at the end is flowed in using PDMS to complete the Field.
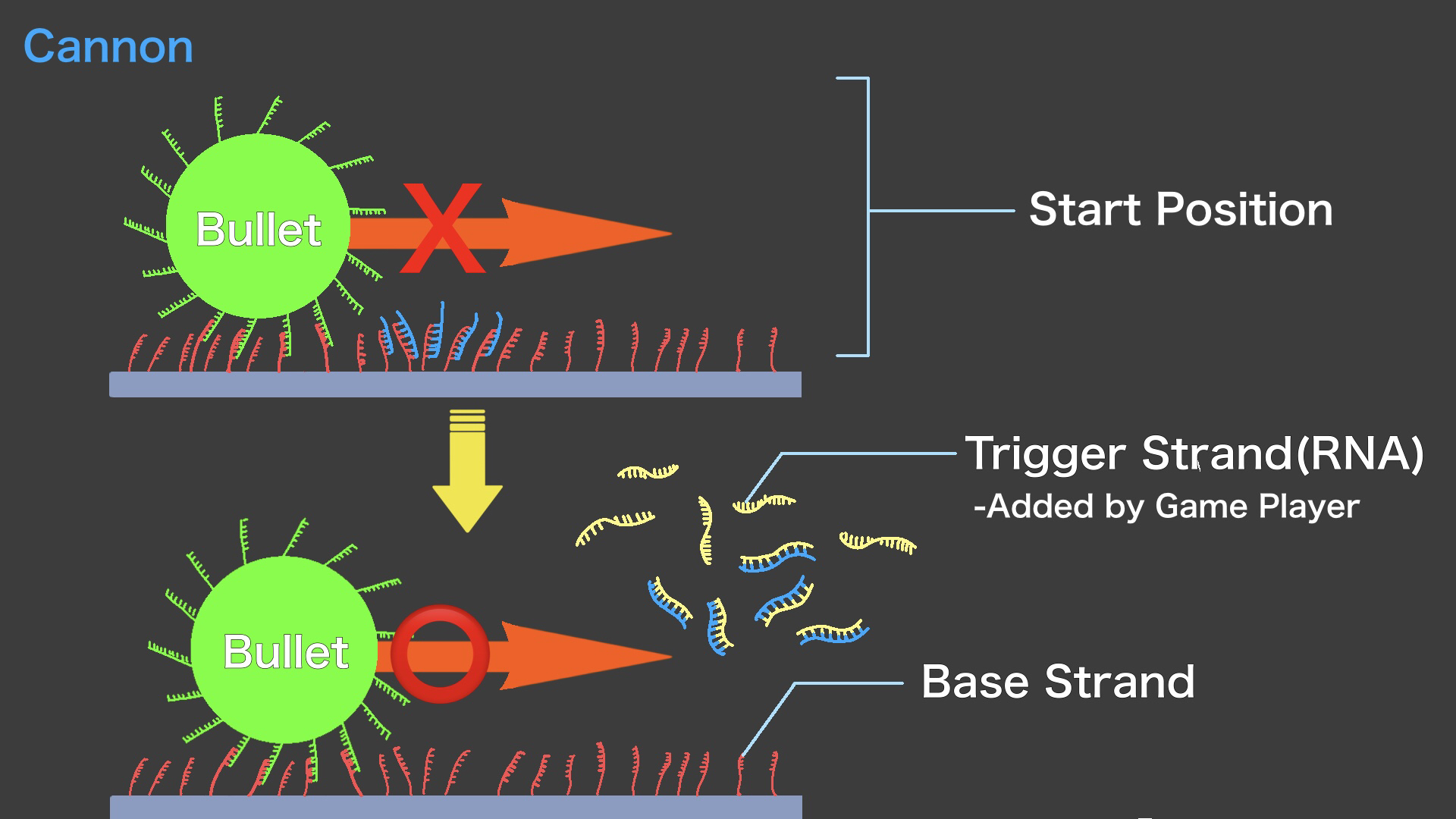
Cannon is composed of two types of RNA strands, called Stopper strand and Trigger strand. Stopper strand is complementary to Base strand, and Trigger strand is complementary to Stopper strand. When the Stopper strands are placed in a part of the Field, those form double-stranded RNA with Base strands. RNase-H does not hydrolyze RNA/RNA double strands, preventing the bullet from advancing beyond that point. The Trigger strand has a sequence that is completely complementary to the Stopper strand, so when the Trigger strand is added to the Field (,where Stopper/Base double strands exist), strand displacement reaction occurs and the Trigger and Stopper strands become a double strand. In other words, this removes the Stopper strand from Base strand. As a result, the Bullet is able to move toward the Enemy. The player of the Micro Invader Game can then drop the Trigger strand onto the Field to fire Bullets. This action is called "Cannon" and represents the firing of Bullets in "Space Invaders".
[1]Guo Liu, et al. (2021) “DNA-Based Artificial Signaling System Mimicking the Dimerization of Receptors for Signal Transduction and Amplification”. Anal. Chem. 2021, 93, 41, 13807-13814
[2]Kevin Yehl, et al. (2016) “High-speed DNA-based rolling motors powered by RNase H”. Nature Nanotechnology 11, 184-190
[3]TIna Karimian, et al. (2022) “Glass Surfaces for PA Simplified and Robust Activation Procedure of Subcellular Micropatterning Experiments”. Biosensors 2022, 12(3), 140